When designing a storage solution for large datasets, the most common families of architectures are data warehouses and data lakes. A data warehouse is a database management system that houses data from multiple data sources. Data is structured in a predefined schema, and can be easily analyzed. A data lake stores all structured, semi-structured and unstructured data in an object storage system. Unlike a data warehouse, it is not necessary to process data in order to add it to a data lake.
While data lakes are often more flexible in terms of writing data when compared with data warehouses, reading data can be more difficult because it has to be validated against a schema. This is known as “schema-on-read”.
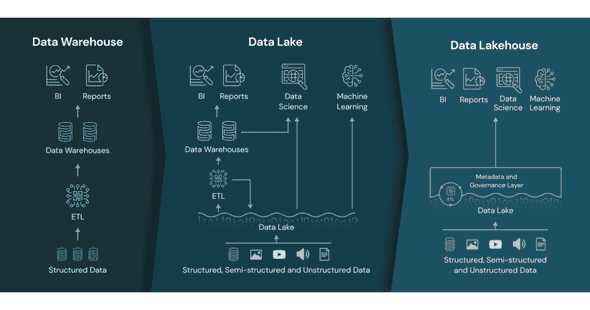
Now consider the following figure, introduced by Databricks:
The left part describes a data architecture using data warehouses, while the middle part is a data architecture combining data warehouses and data lakes. A hybrid data architecture which leverages both data lake and data warehouse architectures can sometimes offer the best of both worlds, but such approaches introduce extra complexity which can make the system more difficult to maintain.
The right part describes a Data Lakehouse architecture. The “lakehouse” architecture, recently introduced by Databricks, aims to provide the best features of both. It builds upon the data lake architecture by introducing a metadata and access layer on top of the usual unstructured object storage layer. This additional layer allows the system to provide the transactional storage of a data warehouse while preserving the flexibility that a data lake offers.
Delta Lake is an open-source project that enables building a data lakehouse1. It runs on top of the existing data lake storage and is fully compatible with Apache Spark APIs. As with most Spark applications, you can interact with data stored in Delta Lake via Python scripts using pyspark, with Spark SQL, in Scala or Java programs using the Delta Lake Scala / Java libraries, or in an interactive Spark shell (Scala or Python). While these approaches enable a number of common use cases, they fall short for users seeking stronger compile-time guarantees.
This is where Sparkle, a Haskell library for writing Spark applications, comes into play. Sparkle was first released in 2016 and aims to provide Spark users with stronger compile-time guarantees as well as Haskell’s concise syntax. We introduced it in a blog post in 2016. In this blog post, we will demonstrate how to use Sparkle with Delta Lake. As an example, we will define and run a series of genomics computations.
How Sparkle Works With Delta Lake
As a first step, let’s use Sparkle to create a simple program which leverages Delta Lake. Generally speaking, Sparkle is an open-source tool for creating self-contained Spark applications in Haskell. It creates JAR files that Spark applications need, and then submits the JAR files to a local or cluster deployment of Spark.
We will perform the following three steps and add the necessary configurations to make Sparkle work with Delta Lake.
- Create an application in the
apps/folder, in-repo or as a submodule. - Build the application.
- Submit the application to a local or cluster deployment of Spark.
1. Create
We start by creating a hello-deltalake Spark application in the apps/ folder.
{-# LANGUAGE OverloadedStrings #-}
module Main where
import Control.Distributed.Spark
import qualified Control.Distributed.Spark.SQL.Dataset as Dataset
main :: IO()
main = forwardUnhandledExceptionsToSpark $ do
-- Set up
conf <- newSparkConf "Hello Deltalake in Sparkle!"
confSet conf "spark.sql.extensions" "io.delta.sql.DeltaSparkSessionExtension"
confSet conf "spark.sql.catalog.spark_catalog" "org.apache.spark.sql.delta.catalog.DeltaCatalog"
session <- builder >>= (`config` conf) >>= getOrCreate
-- Create a table
df1 <- Dataset.range 0 5 1 1 session
Dataset.write df1
>>= Dataset.formatWriter "delta"
>>= Dataset.save "delta-table"
-- Read a table
Dataset.read session
>>= Dataset.formatReader "delta"
>>= Dataset.load "delta-table"
>>= Dataset.show
-- Update a table (overwrite)
df2 <- Dataset.range 5 10 1 1 session
Dataset.write df2
>>= Dataset.formatWriter "delta"
>>= Dataset.modeWriter "overwrite"
>>= Dataset.save "delta-table"
Dataset.read session
>>= Dataset.formatReader "delta"
>>= Dataset.load "delta-table"
>>= Dataset.show
-- Read older versions of data using time travel
Dataset.read session
>>= Dataset.formatReader "delta"
>>= Dataset.optionReader "versionAsOf" "0"
>>= Dataset.load "delta-table"
>>= Dataset.showHere, we created a Spark Session session as an entry point to Spark to work with a data set, and configured it with Delta Lake.
Then, as a simple example, we created a data set df1 as a range 0 1 2 3 4 and wrote it out in the delta format.
Then, we read the data by specifying the path to the folder "delta-table", once again making df1 available as defined.
Next, we ran a batch job to overwrite the data in the data set with df2 - a range 5 6 7 8 9 - by writing it out again to the same delta files.
Right now the data stored as "delta-table" should be the range 5 6 7 8 9.
Lastly, we queried previous snapshots of the Delta table using time travel.
The last line of the code should display the previous data 0 1 2 3 4 as it was before being overwritten with 5 6 7 8 9.
2. Build
We shall use Bazel to build the project. Since we need one build file per directory for it to work, we include the following in a BUILD.bazel file next to the source code to specify all targets for this hello-deltalake Spark application.
Apart from the Sparkle packages, we also declare the Delta Lake binaries from the Maven Central Repository as a dependency of haskell_library.
This allows the main function in HelloDeltalake.hs to be invoked by Spark and Delta Lake.
package(default_visibility = ["//visibility:public"])
load(
"@rules_haskell//haskell:defs.bzl",
"haskell_library",
)
load("@//:sparkle.bzl", "sparkle_package")
haskell_library(
name = "hello-deltalake",
srcs = ["HelloDeltalake.hs"],
deps = [
"//:sparkle-lib",
"@stackage//:base",
"@stackage//:distributed-closure",
"@maven//:io_delta_delta_core_2_11",
],
)
sparkle_package(
name = "sparkle-example-hello-deltalake",
src = ":hello-deltalake",
)Bazel is then invoked to build a JAR file for deployment. We use Nix to isolate the build environment:
$ nix-shell --pure --run "bazel build //apps/hello-deltalake:sparkle-example-hello-deltalake_deploy.jar"3. Submit
The following command runs the application locally with spark-submit in a Nix shell:
$ nix-shell --pure --run "bazel run spark-submit -- --packages io.delta:delta-core_2.11:0.4.0 $PWD/bazel-bin/apps/hello-deltalake/sparkle-example-hello-deltalake_deploy.jar"A glance into the bazel-bin/spark-submit.runfiles/io_tweag_sparkle/ folder will reveal the presence of a sub-folder delta-table, containing versioned parquet files, and another sub-folder _delta_log (also within delta-table), in which transaction logs are automatically written after each commit as JSON files.
Genome-Wide Association Study, From Haskell
Now that we’re able to interact with delta files from Sparkle, we can take a look at a more complex, real-world example from the life sciences: ingesting some genomics data into our delta lake and performing a genome-wide association study against it.
A Genome-Wide Association Study (GWAS) is a study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a phenotype. A phenotype, which is also called a trait, is any observable characteristic of an individual. Phenotypes can be continuous (e.g. height or weight) or discrete (e.g. the presence or absence of a disease). In Genome-Wide Association Studies, linear or logistic regression is a standard approach to identifying genetic variants associated with a continuous or discrete phenotype. To control for confounding associations between genotype and phenotype, GWAS analyses typically take into account a set of covariates, such as age and sex.
In our example, we will first ingest genotypes.vcf – a VCF file containing genetic variants – into Delta Lake, and two CSV files containing phenotype and covariate data into Apache Spark datasets.
Then, we will format the datasets and do the GWAS analysis using Sparkle.
A VCF file contains a header that provides metadata describing the body of the file, and data lines which are unlimited tab-separated lists, each containing information about a position in genome.
To load .vcf files into an Apache Spark dataframe/dataset just as any other file format that Apache Spark supports, we import Glow - an open-source toolkit for working with genomic data, built on Apache Spark and Delta Lake - in Sparkle, and do the GWAS analysis with the built-in Glow functions.
To run the Spark application from Sparkle, we still need to perform the following three steps, which we will go through in detail.
1. Create
In the apps/ folder, we create our deltalake-glow sub-folder where we start to create our Spark application.
The first thing to do is to define a Spark session called session, which is the entry point of the application to interface with data sources and perform activities like reading and writing data.
While creating the Spark session, we set some Spark configurations via confSet to make sure that Delta Lake and Glow packages are loaded.
To access the Glow functions, we need to register them with the Spark session using the registerGlow function.
Additionally, since the GWAS analysis function in Glow requires the covariates in org.apache.spark.ml.linalg.Matrix format, we also need to register the registerUDFDenseMatrix function with the Spark Session, to be able to apply it later.
{-# LANGUAGE OverloadedStrings #-}
{-# LANGUAGE DataKinds #-}
module Main where
import Control.Distributed.Spark
import qualified Control.Distributed.Spark.SQL.Dataset as Dataset
main :: IO ()
main = forwardUnhandledExceptionsToSpark $ do
conf <- newSparkConf "Sparkle Dataset demo"
confSet conf "spark.jars.packages" "io.delta:delta-core_2.11:0.4.0,io.projectglow:glow-spark2_2.11:1.1.2"
confSet conf "spark.hadoop.io.compression.codecs" "io.projectglow.sql.util.BGZFCodec"
confSet conf "spark.sql.extensions" "io.delta.sql.DeltaSparkSessionExtension"
confSet conf "spark.sql.catalog.spark_catalog" "org.apache.spark.sql.delta.catalog.DeltaCatalog"
session <- builder
>>= (`config` conf)
>>= getOrCreate
>>= registerGlow
>>= registerUDFDenseMatrixNext, we ingest the genotype data genotypes.vcf into Delta Lake as an Apache Spark data set dfBaseVariant.
Since the whole schema is too long to display, we will only show the data and schema of the genotypes column.
The data we use here are all test data from the Glow project.
Dataset.read session
>>= Dataset.formatReader "vcf"
>>= Dataset.load "apps/deltalake-glow/genotypes.vcf"
>>= Dataset.write
>>= Dataset.formatWriter "delta"
>>= Dataset.modeWriter "overwrite"
>>= Dataset.save "delta-table-glow"
dfBaseVariant <- Dataset.read session
>>= Dataset.formatReader "delta"
>>= Dataset.load "delta-table-glow"
Dataset.selectDS dfBaseVariant ["genotypes"]
>>= Dataset.show
Dataset.selectDS dfBaseVariant ["genotypes"]
>>= Dataset.printSchemaThe result looks as follows:
+----------------------------+
| genotypes|
+----------------------------+
|[[HG00096, true, [0, 0]],...|
+----------------------------+
root
|-- genotypes: array (nullable = true)
| |-- element: struct (containsNull = true)
| | |-- sampleId: string (nullable = true)
| | |-- phased: boolean (nullable = true)
| | |-- calls: array (nullable = true)
| | | |-- element: integer (containsNull = true)Before we apply the GWAS analysis to our data, we need to do some data cleaning. This involves:
- getting the number of alternate alleles for an array of
genotypestructs using thegenotypeStatesfunction (see here for more details); - ingesting phenotype data into Apache datasets and converting the first trait
Trait_1to an array of doubles; - ingesting covariate data into Apache datasets and turning these into a matrix; and finally
- combining these three variables into one single data set
dfVariantPhenoCov.
dfVariant <- Dataset.col dfBaseVariant "genotypes"
>>= genotypeStates
>>= \colGenotypeStates
-> Dataset.withColumn "genotype values" colGenotypeStates dfBaseVariant
dfPhenotype <- Dataset.read session
>>= Dataset.formatReader "csv"
>>= Dataset.optionReader "header" "true"
>>= Dataset.optionReader "inferSchema" "true"
>>= Dataset.load "apps/deltalake-glow/continuous-phenotypes.csv"
dfPhenColNames <- Dataset.columns dfPhenotype
phTrait1 <- Dataset.selectDS dfPhenotype [dfPhenColNames !! 1]
dfVariantPheno <- Dataset.as double phTrait1
>>= Dataset.collectAsList
>>= lit
>>= \phTrait1Col
-> Dataset.withColumn "phenotype values" phTrait1Col dfVariant
phTrait1NameCol <- lit (dfPhenColNames !! 1)
dfVariantPheno1 <- Dataset.withColumn "phenotype" phTrait1NameCol dfVariantPheno
dfCovariates <- Dataset.read session
>>= Dataset.formatReader "csv"
>>= Dataset.optionReader "header" "true"
>>= Dataset.optionReader "inferSchema" "true"
>>= Dataset.load "apps/deltalake-glow/covariates.csv"
>>= Dataset.drop "sample_id"
nRowsCov <- Dataset.count dfCovariates
covColNames <- Dataset.columns dfCovariates
let nRows = (fromIntegral nRowsCov) :: Int32
let nCols = (fromIntegral (Prelude.length covColNames)) :: Int32
dfVariantPhenoCov <- concatCov columnAsDoubleList dfCovariates covColNames
>>= lit
>>= \covariateCol
-> Dataset.withColumn "covariates" covariateCol dfVariantPheno1
>>= \dfList
-> callUDFDenseMatrix dfList nRows nCols "covariates"Now we have done all the data formatting jobs and have the data set dfVariantPhenoCov, which we will hand to the GWAS analysis function later. We print the schema of dfVariantPhenoCov here.
Dataset.selectDS dfVariantPhenoCov ["genotype values", "phenotype values", "cov"]
>>= Dataset.printSchemaWe can see that the schema of genotypes (column genotype values), phenotypes (column phenotype values), and covariates (column cov) of dfVariantPhenoCov are as below:
root
|-- genotype values: array (nullable = true)
| |-- element: integer (containsNull = true)
|-- phenotype values: array (nullable = false)
| |-- element: double (containsNull = true)
|-- cov: matrix (nullable = true)Finally, we hand the dataset to the linearRegressionGwas function, and insert the result resultExpand into our Delta Lake.
colGeno <- Dataset.col dfVariantPhenoCov "genotype values"
colPheno <- Dataset.col dfVariantPhenoCov "phenotype values"
colCov <- Dataset.col dfVariantPhenoCov "cov"
colContig <- Dataset.col dfVariantPhenoCov "contigName"
colStart <- Dataset.col dfVariantPhenoCov "start"
colPhenoName <- Dataset.col dfVariantPhenoCov "phenotype"
colRegression <- linearRegressionGwas colGeno colPheno colCov
>>= \regressionColumn
-> alias regressionColumn "stats"
result <- Dataset.select dfVariantPhenoCov [colContig, colStart, colPhenoName, colRegression]
resultExpand <- expr "expand_struct(stats)"
>>= \colStats
-> Dataset.select result [colContig, colStart, colPhenoName, colStats]
Dataset.show resultExpand
Dataset.write resultExpand
>>= Dataset.formatWriter "delta"
>>= Dataset.modeWriter "overwrite"
>>= Dataset.save "delta-table-glow-result"
This produces the following result:
+----------+-------+------------------+-----------------+------------------+------------------+
|contigName| start| phenotype| beta| standardError| pValue|
+----------+-------+------------------+-----------------+------------------+------------------+
| 21|9411238|Continuous_Trait_1|0.133214502956773|0.8721198784834174|0.8786093812014514|
+----------+-------+------------------+-----------------+------------------+------------------+2. Build
We include the following in a BUILD.bazel file next to the source code.
Here, we add exports_files so that our program can use these files.
package(default_visibility = ["//visibility:public"])
exports_files(["genotypes.vcf", "continuous-phenotypes.csv", "covariates.csv"])
load(
"@rules_haskell//haskell:defs.bzl",
"haskell_library",
)
load("@//:sparkle.bzl", "sparkle_package")
haskell_library(
name = "deltalake-glow",
srcs = ["DeltalakeGlow.hs"],
deps = [
"//:sparkle-lib",
"@maven//:io_delta_delta_core_2_11",
"@maven//:io_projectglow_glow_spark2_2_11",
"@io_tweag_inline_java//jvm",
"@stackage//:base",
"@stackage//:distributed-closure",
"@stackage//:text",
],
)
sparkle_package(
name = "sparkle-example-deltalake-glow",
src = ":deltalake-glow",
resource_jars = ["@io_tweag_inline_java//jvm-batching:jar"],
)And then we ask Bazel to build a JAR file for deployment:
$ nix-shell --pure --run "bazel build //apps/deltalake-glow:sparkle-example-deltalake-glow_deploy.jar"3. Submit
Finally, we can submit the job using Spark:
$ nix-shell --pure --run "bazel run spark-submit-with-data -- --packages io.delta:delta-core_2.11:0.4.0,io.projectglow:glow-spark2_2.11:1.1.2 $PWD/bazel-bin/apps/deltalake-glow/sparkle-example-deltalake-glow_deploy.jar"Similarly to the former hello-deltalake example, the genotype data we ingested into our Delta Lake will be found in bazel-bin/spark-submit.runfiles/io_tweag_sparkle/delta-table-glow/ and the result will be found in bazel-bin/spark-submit.runfiles/io_tweag_sparkle/delta-table-glow-result/.
Conclusion
In this blog post, we gave an idea of how to build a Delta Lake in Sparkle, by creating a hello-deltalake application doing some basic read/write operations, and another deltalake-glow application running some computations on genomics data.
All the code for these two examples in this blog post is available here.
We hope these examples will help you explore with your own examples in Sparkle and Delta Lake.
- Historically, portions of Delta Lake were closed source, but with version 2.0 the entirety of Delta Lake has been open-sourced.↩
Behind the scenes
Zhihan is a data scientist/engineer with expertise in Machine Learning. Holding a PhD in Geometric Topology and Probabilities, she transitioned from a pure mathematics background to apply her knowledge in industry. Currently at Tweag, Zhihan focuses on developing data engineering solutions and cloud deployment. Beyond her technical skills, she has a passion for music, arts, literature, and cooking.
If you enjoyed this article, you might be interested in joining the Tweag team.