Artificial Intelligence (AI). Machine Learning (ML). Deep Learning. Neural Networks (NNs). Large Language Models (LLMs)… The list of hyped buzzwords goes on and on, even more so since ChatGPT made a wider audience realize what is now achievable. As scary or awe-inspiring as it is, one can’t deny the great impact AI can have when applied to fields with positive social value, such as healthcare.
The MedTech industry is buzzing thanks to a continuous stream of innovation, promising to be more precise, efficient and accessible than ever. In particular oncology, a branch of medicine that focuses on cancer, could benefit immensely from these new technologies, which may enable clinicians to detect cancer earlier and increase chances of survival. Detecting cancerous cells in microscopic photography of cells (Whole Slide Images, aka WSIs) is usually done with segmentation algorithms, which NNs are very good at. While using ML and NNs for image segmentation is a fairly standard task with established solutions, doing it on WSIs is a different kettle of fish. Most training pipelines and systems are designed to handle fairly small, sub-megapixel images. In the case of WSIs, the image is so huge that a single file is at least a few hundred megabytes and can be dozens of gigabytes. To allow innovation in medical imaging with AI, we need efficient and affordable ways to store and process these WSIs at scale.
In this blog post, I will explain the underlying technical challenges and share the solution that we helped implement at kaiko.ai, a MedTech startup in Amsterdam that is building a Data Platform to support AI research in hospitals.
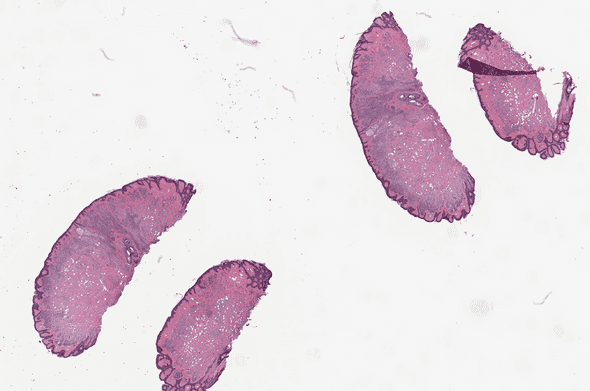
A bunch of Whole Slide Images
Whole Slide Images (WSIs) are ubiquitous in digital pathology. These files store microscopic photography images of a slide, a piece of glass with cells on it.
OpenSlide test data: CMU-1.tiff
Since the capture is done through a microscope, an image of a few centimeters becomes millions of pixel long. The ratio from pixels to real distance is called micrometer per pixel (aka MPP). The lower the MPP, the more the image is zoomed in.
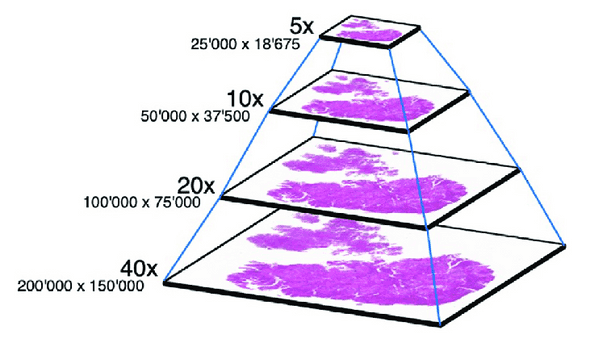
An image of a slide with a low MPP is very large thus slow to read, which is not fit for every use case. For example, someone might just need to visually confirm the quality of the overall image at a higher MPP. To allow using an MPP that fits any usage, multiple images at higher MPP are stored as well, like a pyramid.
A pyramid of images, from “Multi_Scale_Tools: A Python Library to Exploit Multi-Scale Whole Slide Images”, N. Marini et al
This results in a very large amount of data for a single slide, often a few gigabytes per slide, which is all stored in one big file. A single hospital makes many captures a day, producing terabytes of such data to store and process.
To store this data, hospitals are often equipped with on-premises infrastructure, more or less provided by the same manufacturer of the capture devices. These decades-old systems were tailored to support doctors in their traditional tasks, like displaying a WSI for manual analysis. But the rise of Machine Learning in research has driven a need for new systems that are more performant and more flexible.
Thankfully, cloud-based infrastructure is now an established solution which can help do this in a cost-effective way. As a simple solution, files can be stored on cloud storage services, such as Azure Blob Storage or AWS S3, which can scale more easily than on-premises infrastructure. However, it is a big shift in architecture that leads to numerous technical challenges.
Reading WSIs from Blob Storage
The first basic challenge is to actually read the image. Whether displaying it on a screen or feeding it to a neural network, it is fundamental to have a tool to turn the stored bytes into a meaningful representation. Fortunately, there is OpenSlide, the most widely used open-source library to read WSI files… or so we’d like. But as it turns out, we can’t use it.
Although it has Python bindings, OpenSlide is implemented in C and reads files using standard OS file handlers, however our data sits on cloud storage that is accessible via HTTP. This means that, to open a WSI file, one needs to first download the entire file to disk, and only then can they load it with OpenSlide. But then, what if we need to read tens of thousands of WSIs, a few gigabytes each? This can total more than what a single disk can contain. Besides, even if we mounted multiple disks, the cost and time it would take to transfer all this data on every new machine would be too much. In addition to that, most of the time only a fraction of the entire WSI is of interest, so downloading the entire data is inefficient.
A solution is to read the bytes that we need when we need them directly from Blob Storage.
fsspec is a Python package that allows us to define “abstract” filesystems, with a custom implementation to list, read and write files.
One such implementation, adlfs, works for Azure Blob Storage.
Thanks to these libraries, we can keep the data on cloud storage and still read it partially.
from adlfs import AzureBlobFileSystem
# anon=False to use local azure-cli credentials
fs = AzureBlobFileSystem(anon=False, account_name="my_account")
with fs.open("container/file.svs") as f:
# read the first 256 bytes
print(f.read(256))Written on top of fsspec, tiffslide is another Python package that is supposed to be a drop-in replacement of openslide-python, the Python bindings of OpenSlide.
Since it uses fsspec behind the scenes, it can be used to directly read the WSIs that are stored on Azure Blob Storage without copying them first to disk.
From our experience, the performance is somewhat acceptable for our machine learning use cases, thanks to the fast connection in cloud computing data centers.
Another option would be to use blobfuse, which uses FUSE to make it seem like the Azure Blob Storage container is mounted just like any disk.
Unfortunately, we have found this solution to be quite limited, as it needs to download the entire file locally which gives a huge overhead.
There is a “streaming” mode which is supposed to directly read byte ranges from Blob Storage, but we did not investigate further because blobfuse only works on Linux, while our users needed it to run on MacOS.1
Patching on-the-fly
The most common way to train NNs is using Stochastic Gradient Descent (or similar). This means we randomly loop over samples and use backpropagation to “train” the model. In this loop, one would usually want to iterate on batches of samples. So at every step, instead of computing the gradient on a single sample, it is computed on multiple samples.2 This supposedly makes the gradient descent more stable and epochs faster. Usually, the batch size ranges from tens to hundreds of samples.
As stated earlier, WSIs are quite large, which means that it would be hard to stack so many of them in RAM, let alone on the GPU.
We are not interested in the entire image either.
In oncology, WSIs are cut down to smaller images call patches, which are of a more reasonable size.
To generate a patch, one needs to read a region of the image, for instance using openslide.OpenSlide.read_region or tiffslide’s equivalent.
We can either generate all patch images and store them, or we can patch “on-the-fly”. From our discussions with researchers, precomputing patches usually takes a lot of time and is quite inflexible. In fact, one could look at parameters such as the patch size as a hyper-parameter to the model which should be tuned. Since it takes a lot of time to generate all the patches of all WSIs, this creates a very long feedback loop. This also multiplies the storage cost, as the weight of all patch images is roughly the same as the WSIs. For these reasons, we tried to implement patching “on-the-fly”, or online patching.
The minimalistic script below shows how we can do online patching to train a PyTorch model.
import pytorch_lightning as pl
from pydantic.dataclasses import dataclass
from tiffslide import TiffSlide
from torch.utils.data import DataLoader
@dataclass(frozen=True)
class PatchSpec:
level: int
x: int
y: int
width: int
height: int
class PatchDataset(data.IterableDataset):
def __init__(self, slides_specs: dict[str, list[PatchSpec]]):
self._slides_specs = slides_specs
def __iter__(self):
for file_uri, specs in self._slides_specs.items():
with fsspec.open(file_uri) as f:
slide = TiffSlide(f)
for spec in specs:
yield slide.read_region(
location=(spec.x, spec.y),
level=spec.level,
size=(spec.width, spec.height),
)
if __name__ == "__main__":
model: pl.LightningModule = ... # load some pytorch NN model
slides_specs: dict[str, list[PatchSpec]] = ... # load training metadata
dataset = PatchDataset(slides_specs=slides_specs)
train_loader = DataLoader(dataset)
trainer = pl.Trainer(limit_train_batches=100, max_epochs=1)
trainer.fit(model=model, train_dataloaders=train_loader)
...Unfortunately, we found that loading each patch individually into batches like this is slower than the time it takes to run a training iteration step.
What happens is:
- a batch of patches is generated and fed to the training loop;
- while it does a training step (inference + backprop + weight update), it generates the next batch of patches;
- unfortunately, the training step is finished before it has generated the next batch of patches, so the GPU is idle for a while.
This leads to low usage of the GPU and slower training, which is not ideal.
Ideally, we would meet the following constraint:
ttrain_step>nbatch_size×tload_patch
Where:
- ttrain_step is the average time the GPU takes to run a training step;
- nbatch_size is the batch size;
- tload_patch is the average time it takes to load a single patch.
We don’t really want to change the batch size, and we can hardly improve ttrain_step. So in order to speed up the training and reach 100% GPU utilization, we need to generate patches faster.
… or we can change the equation!
ttrain_step>nparallelnbatch_size×tload_patch3
Where nparallel is the number of parallel steps in the data loading.
We can easily increase parallelism thanks to torch.utils.data.DataLoader’s num_workers parameter: setting it to a value above 1 makes it load batch items in parallel.
This requires some changes to the above implementation of PatchDataset, but nothing too fancy.
Unfortunately, we witnessed that the amount of parallelism required to meet the constraint was way above the number of CPU cores on the machines. Threads would quickly clutter and the throughput was limited.
But you know what they say: if one machine is not enough, use more machines!
Distributed data loading
Fortunately, we can scale horizontally by distributing over multiple machines. This is where Ray comes into play.
Ray is “an open-source unified compute framework that makes it easy to scale AI and Python workloads”.4
One of Ray’s selling points is how simple it is to go from a local environment, to develop and debug, to a production environment at scale.
Much like Spark, its module ray.data is focused on loading and processing data as scale.
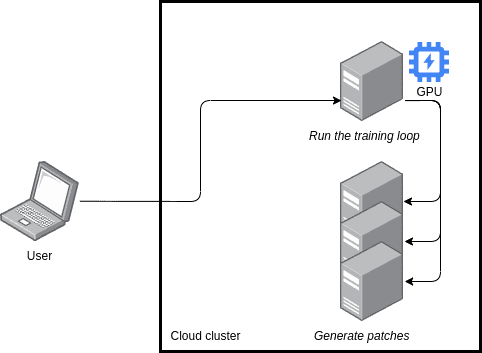
With Ray, we could use the following architecture.
A researcher is working on their laptop, which connects to a VM in a cloud cluster. This VM is the one on which the training loop happens, so it has a beefy GPU, and it can connect to other VMs in the cluster to distribute the workload on patching.
This allows us to increase nparallel as much as we want, if we want to optimize for utilization, until it meets the constraints.
Below, we demonstrate a simplified way of using Ray to distribute the WSI processing to feed the training loop.
import pandas as pd
import PIL.Image
import ray.data
import torch.utils.data
...
def read_patch(record) -> PIL.Image.Image:
file_uri: str = record["file_uri"]
spec: PatchSpec = record["spec"]
with fsspec.open(file_uri) as f:
slide = TiffSlide(f)
return {
"patch": slide.read_region(
location=(spec.x, spec.y),
level=spec.level,
size=(spec.width, spec.height),
),
}
def get_data_loader(
slides_specs: dict[str, list[PatchSpec]],
batch_size: int,
prefetch_blocks: int,
) -> torch.utils.data.IterableDataset:
df = pd.DataFrame({"file_uri": k, "spec": v} for k, v in slides_specs.items())
ds = (
ray.data.from_pandas(df)
# parallelize into blocks of size `batch_size`
.repartition(num_blocks=len(df)/batch_size)
# turn into a dataset pipeline to evaluate blocks lazily
.window(blocks_per_window=1)
# read patches
.map(read_patch)
)
return ds.to_torch(
feature_columns="patch",
batch_size=batch_size,
prefetch_blocks=prefetch_blocks,
)This snippet can benefit from numerous optimizations and improvements. However the rough idea is as follows:
- We flatten and turn the previous
slides_specsinto aray.data.Datasetto work with Ray. .repartitiondistributes it into many blocks,5 otherwise.mapwould compute everything on the same worker..windowturns the dataset into aray.data.DatasetPipeline, otherwise.mapwould compute all blocks.- Finally,
.mapmakes a pipeline that will effectively read the WSI patches when iterated.
Then this dataset can be plugged to our PyTorch script using .to_torch.
However this code can’t be run locally, as a single computer will not have enough cores to parallelize it enough.
Reach for the clouds
In order to scale this distributed processing pipeline, we ought to use the cloud!
In our case, we decided to use AKS, Azure’s managed Kubernetes, to set up the compute cluster,
and kuberay, an open source toolkit to run Ray applications on Kubernetes, to set up the ray clusters on top of it.
More specifically, kuberay will deploy the Custom Resource Definitions, Operator and Service that make it easy to manage clusters with YAML files and a CLI.
Since we use OpenSlide and other specific packages, we need pods to run on our own Docker images that will have our tools and libraries installed. Thus we connect AKS to an Azure Container Registry in our Virtual Private Cloud (VPC).
Now, let’s imagine we have the following Ray cluster in our Kubernetes’ default namespace:
- 1 head pod, GPU, named
ray-head - 8 worker pods, no GPU, named
ray-worker-{number},{number}ranging from1to8
Once kubectl is configured on the user’s laptop, they can SSH into the head worker with the following command.
$ kubectl attach ray-headThis allows the user to work directly in the pod from their terminal.
However, it is usually preferable to use Ray’s job system instead.
We can send a job to the ray cluster thanks to Ray’s CLI,
but that requires communicating with the Ray cluster’s dashboard server.
It possible to do this with kubectl port-forward.
$ kubectl port-forward ray-head 8265:8265 &
$ ray job submit --runtime-env-json '{"py_modules":["mymodule"]}' -- python -m mymoduleThe runtime environment allows us to easily specify a local Python module that we are working on, here mymodule, that should be pushed from the user’s laptop to the cluster, among many other settings.
With this setup, a user can easily write a Python module, run it locally to debug it, then run it at scale on a powerful cluster.
Conclusion
All in all, it is about building a platform for researchers to focus on what they really want to do: research. Machine Learning researchers should write their ML model and data processing, push that to a service, and it should just work. We achieved that to a certain degree thanks to Ray and Kubernetes.
In our case, processing the data “online” at each training iteration was the bottleneck, and doing it all “offline” was not a good solution. Fortunately this workload could be parallelized. But one computer wasn’t enough, so we used distribution.
This is why Ray was such a good fit: it provided an easy way to write distributed code in Python, the ability to schedule and run jobs on Kubernetes, and also a convenient CLI to submit these jobs.
Many challenges still remain. Ray doesn’t have a proper job queue that can schedule jobs when resources are available, so teams have to check the availability of compute on the cluster themselves before they can submit a job. Related to that, it is not possible to control and assign resources to teams; unlike Slurm, which is widely used in the academic world, but is less flexible.
Our work built the core of a Data Platform which achieves flexibility with reasonable usability, suited for our client’s use case and all its various ad-hoc requirements that come from working on diverse and unstructured data.
- An issue is open to handle this case, but it made us decide not to use it. See blobfuse#986: “Build on MacOS”↩
- To learn more about Stochastic Gradient Descent and why we use batches, check out “Batch, Mini Batch & Stochastic Gradient Descent”↩
- This equation here is an approximation. Indeed, parallisation does not always lead to a linear speed-up (see Amdahl’s law).↩
- Cited from its official web page↩
- A “block” is a core concept in Ray Data. Basically it is a set of rows that is always manipulated as a whole. Concretely, to process a dataset on two different workers at the same time, you need said dataset to be repartitioned on at least two blocks. To learn more about it, see Ray’s Dataset documentation.↩
Behind the scenes
Guillaume is a versatile engineer based in Paris, with fluency in machine learning, data engineering, web development and functional programming.
If you enjoyed this article, you might be interested in joining the Tweag team.